Varies of microscopy methods were put forwarded to investigate the biological problems in different spatial and temporal scales. However, each imaging method can either achieve high spatial or temporal resolution. None of them can achieve milliseconds temporal resolution and sub-diffraction spatial resolution in 3 dimensions at the same time. On the other hand, many biological interactions happen in the milliseconds time scale and tens of nanometers spatial scale, like DNA transcription, protein folding, et. al.
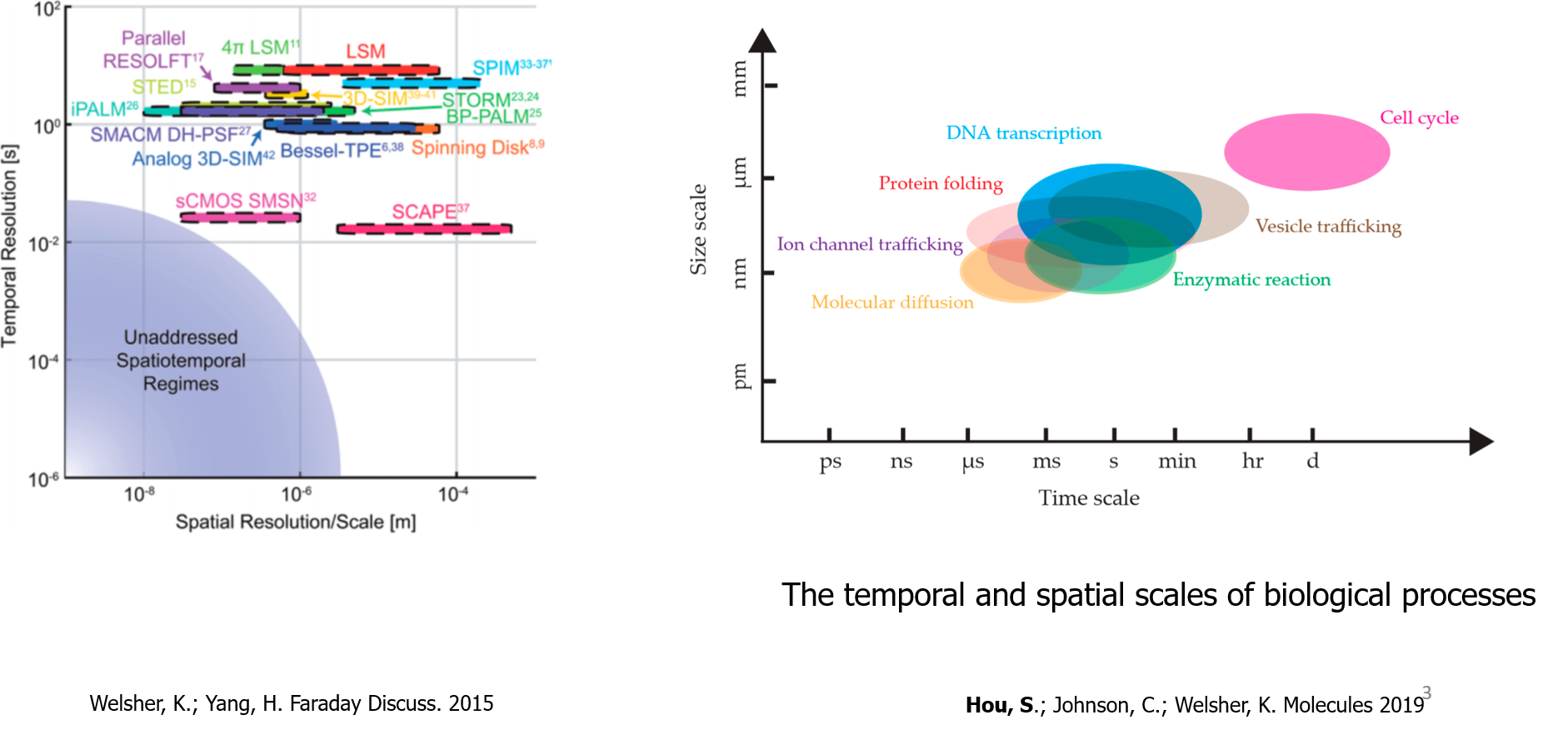
Let's check the relations among field of view, temporal resolution, and spatial resolution. There are always tradeoff relations among them. They are confined by the limited photons, sampling frequency, and illumination time. For example, between the temporal and spatial resolution, because of the limited photons, increase the temporal resolution means there are fewer photons in each frame, which will result in the decrease of spatial resolution. The size of the field of view also affects the illumination time and spatial sampling frequency, which will further affect the temporal and spatial resolution.
Single particle tracking (SPT) aims to obtain the position of the particle with a time sequence, which is called the trajectory of the particle. In traditional SPT, a series of images are acquired and the positions of particles are identified in these images, and finally, particle locations are connected to form trajectories. In these methods, usually wide-field illumination is used. Therefore, the temporal resolution is determined solely by the imaging rate and the imaging depth also is limited.
Real-time 3D single particle tracking microscopy (RT-3D-SPT) is different with traditional single particle tracking methods. In RT-3D-SPT, instead of taking images of particles with the camera, it aims to use real-time information to lock-on to a moving target using active feedback. RT-3D-SPT only follows a particle of interest, which endows it a large axial imaging range and high spatiotemporal resolution. If the active feedback can be made sensitive and fast enough, it can be used to apply SPC-APD based single-molecule detection to capture the dynamics of single molecules in varying environments, such as the cytoplasm of living cells, over long times with high temporal precision.
3D Dynamic Photon Localization Tracking (3D-DyPLoT)
We developed the 3D-DyPLoT by using a tunable acoustic gradient (TAG) lens and a 2D electro-optic deflector (2D-EOD) to dynamically drive a single focused laser spot around the focal volume in 3D (Hou et al., Optics Letters, 2017; Hou and Welsher, J. Vis. Exp., 2018).
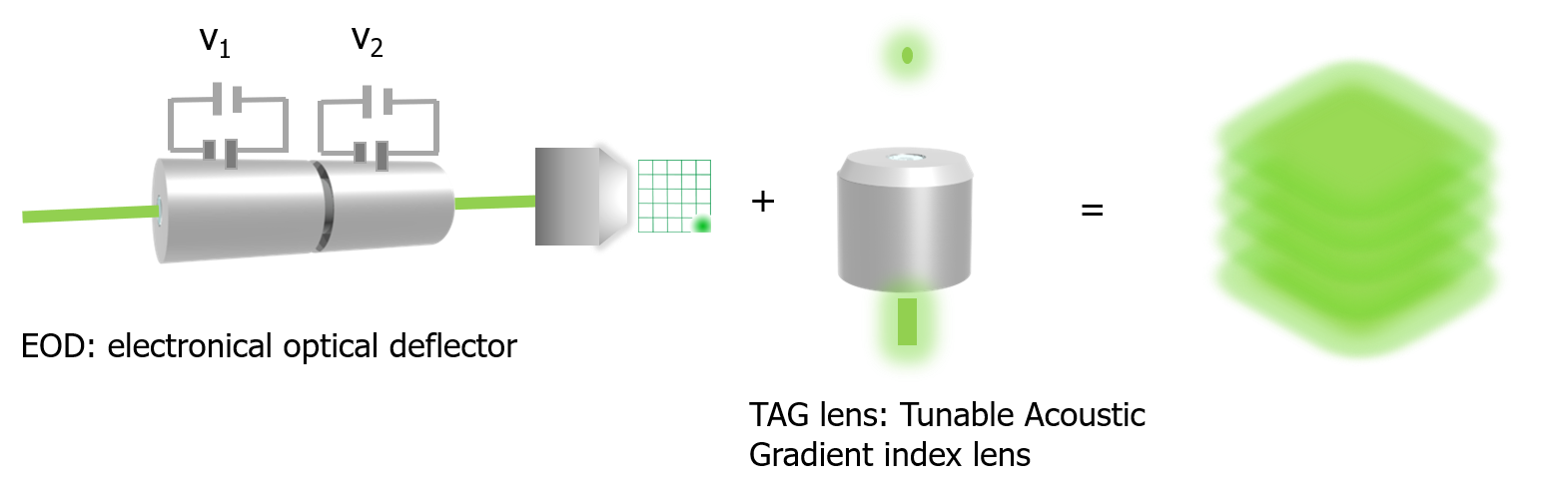
Schematic of 3D-DyPLoT
Utilizing an optimized position estimation algorithm, 3D-DyPLoT can track single particles with high tracking speed (up to 20 μm2/s) and high localization precision at photon count rates as low as 10 kHz. Importantly, the large effective detection area allows the system to easily pick up fast-moving particles, while still demonstrating high precision localization (σx = 6.6 nm, σy = 8.7 nm, σz = 15.6 nm).
Real-time 3D tracking of freely diffusing 100 nm nanoparticles using 3D Dynamic Photon Localization Tracking (3D-DyPLoT). A single freely diffusing particle is “locked” in the focus of the objective lens using a fast piezoelectric stage and an optical feedback loop.
The 3D-DyPLoT shows the best overall performance in the field (Hou et al., Molecules, 2019). Overall, 3D-DyPLoT provides a fast and robust method for real-time 3D tracking of fast and lowly emitting particles, paving the way to more widespread application to relevant biological problems.
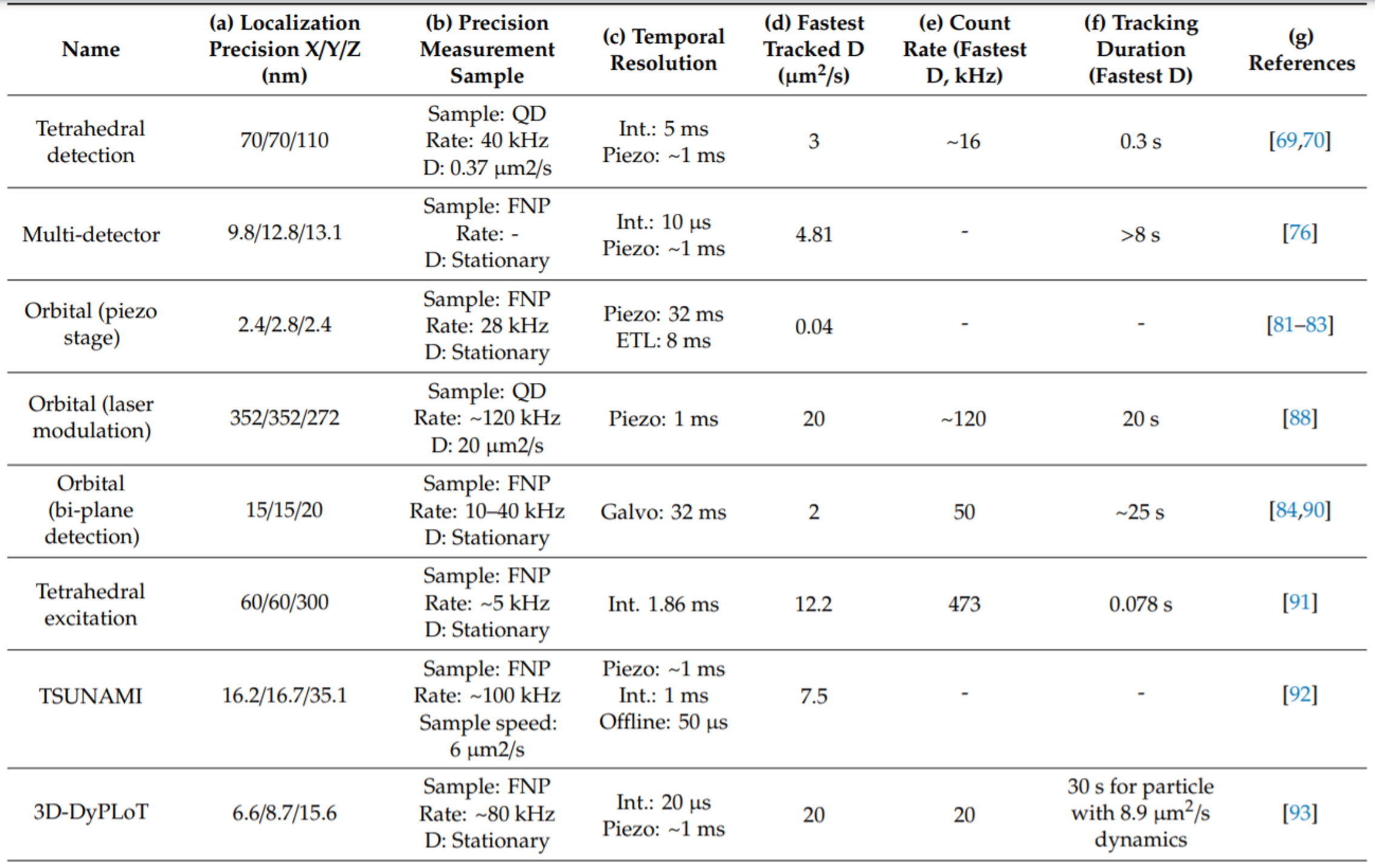
Comparison of 3D-DyPLoT with other real-time 3D single particle tracking methods
3D Single-Molecule Active Real-time Tracking (3D-SMART)
The current single-molecule imaging methods are limited by their observation time and confined environment. To overcome these limitations, we developed the 3D-SMART (Hou et al., Nature Communications, 2020). The 3D-SMART is capable of continuously tracking single fluorophore molecules for several minutes at a time with the photon-limited temporal resolution, two orders of magnitudes longer than previously reported methods. As a demonstration, 3D-SMART was applied to actively track single Atto 647N fluorophores in 90% glycerol solution with an average duration of ~16 s at count rates of ~10 kHz.3D-SMART represents a critical step towards the untethering of single-molecule spectroscopy and promises to be a powerful tool for capturing the dynamics of single biomolecules at high speeds and over 3D distances.

Real-time measurement of transcription on a single, freely-diffusing dsDNA. The top panel shows the dsDNA molecule’s real-time 3D trajectory. The trajectory is labelled with a green sphere when mRNA (fastFISH) signal is observed. The bottom panels show the intensity of Atto 647N labeled dsDNA (red) and the readout of the ssDNA (green). The real-time photon readout at the individual XY laser scan positions is displayed in the bottom right.[/caption]
3D Precision Adaptive Real-time Tracking (3D-PART)
To capture heterogeneous biological interaction dynamics with high precision, we developed the 3D-PART (Hou and Welsher, Small, 2019). Using a real-time measurement of a rapidly diffusing particle’s positional variance, the 3D-PART microscope adjusts active-feedback parameters to trade tracking speed for precision on demand. The adaptive precision is achieved by modulating the size of the laser focus scanning pattern with the particle’s moving speed.
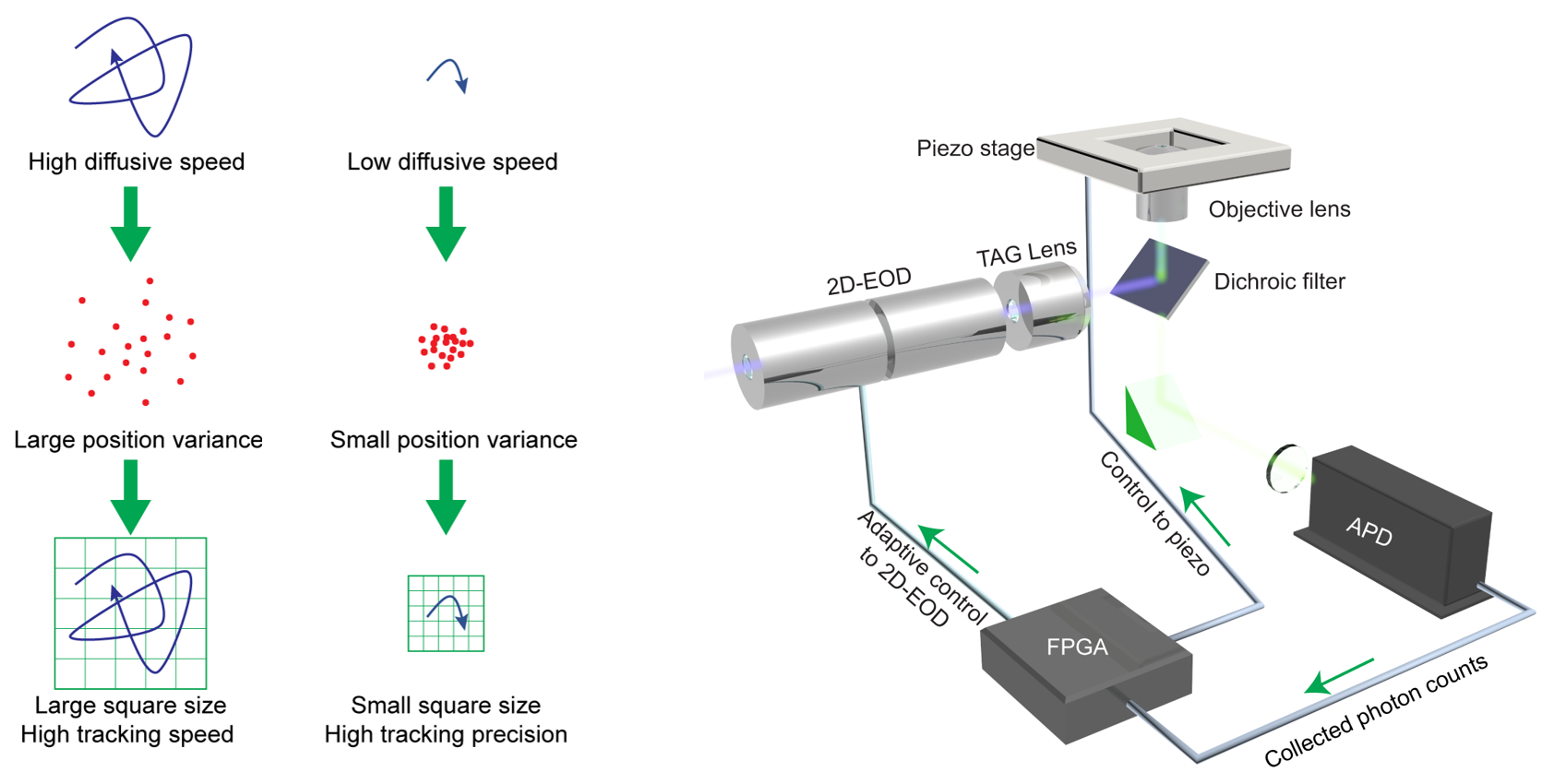
Schematic of 3D-PART
To demonstrate the biological applicability of this adaptive precision tracking method, 3D-PART was then applied to monitoring the viral first contacts of virus-like particles to the surface of live cells, allowing direct and continuous measurement of the viral particle at initial contact with the cell surface.
3D-PART measurement, top-down view, and zoom-in of a single virus-like particle landing on the membrane of a HuH7 cell.